Brief introduction to KisSplice
Introductory slides
KisSplice Docker
KisSplice, KisSPlice2RefGenome and kissDE are also available through a Docker container named kissplice-pipeline. To pull the docker image, use:docker pull dwishsan/kissplice-pipeline
The training dataset contains command lines to use with or without docker.I would like the training on:
The SKNSH datasetThe TALS LCL dataset
KisSplice on SKNSH data
The data
The fastq files can be found herewget ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/data/*fastq
Those correspond to long polyA+ RNAs from SKNSH cell lines. This dataset was generated in the context of the ENCODE project (SRA: SRR315315,SRR315316,SRR534309,SRR534310). There are two experimental conditions (treated with retinoic acid, untreated), and two replicates per condition. Only 10M reads per replicates are analysed here. For an analysis of the full dataset, see hereRunning KisSplice
Here is the command in order to run KisSplice on this dataset :
kissplice -r SknshRACellRep1_10M.fastq -r SknshRACellRep2_10M.fastq
-r SknshCellRep3_10M.fastq -r SknshCellRep4_10M.fastq
mkdir ~/training_Sknsh
wget -P ~/training_Sknsh ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/data/*fastq
docker run --rm -v ~/training_Sknsh:/data dwishsan/kissplice-pipeline kissplice -r /data/SknshRACellRep1_10M.fastq -r /data/SknshRACellRep2_10M.fastq -r /data/SknshCellRep3_10M.fastq -r /data/SknshCellRep4_10M.fastq --counts 0 -o /data/KisSplice
All output files of kissplice are here
Aligning to the reference genome
STAR can be used to align the alternative splicing events found by kissplice back to the reference genome. Here is the reference genome in fasta format : Homo_sapiens.GRCh38.dna_sm.primary_assembly.faAnd the associated gtf : Homo_sapiens.GRCh38.84.gtf
Building STAR index of the genome :
STAR --runMode genomeGenerate --genomeDir STAR_index --genomeFastaFiles Homo_sapiens.GRCh38.dna_sm.primary_assembly.fa --sjdbGTFfile Homo_sapiens.GRCh38.84.gtf
The build index can be found hereAligning to the reference genome :
STARlong --genomeDir STAR_index --readFilesIn results_k41_coherents_type_1.fa
The alignment (in sam format) can be found hereDocker can also be used to run STAR, or any other aligner, with, for instance, the Biocontainers image
Running KisSplice2RefGenome
kissplice2refgenome -a Homo_sapiens.GRCh38.84.gtf --counts 0 Aligned.out.sam
Or, with docker:
wget -P ~/training_Sknsh ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/STAR_asEventAlign/Aligned.out.sam
wget -P ~/training_Sknsh ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/annotation/Homo_sapiens.GRCh38.84.gtf
docker run --rm -v ~/training_Sknsh:/data dwishsan/kissplice-pipeline kissplice2refgenome -a /data/Homo_sapiens.GRCh38.84.gtf -o /data/training_Sknsh_k2rg_Type_1 --counts 0 /data/Aligned.out.sam
The ouput file can be found here : k2rg-v1.0.1_sknsh.txt
Running kissDE
The last version of kissDE can be downloaded here.
#!/usr/bin/Rscript
library(kissDE)
countsData<-kissplice2counts("k2rg-v1.0.1_sknsh.txt", k2rg=TRUE)
conditions<-c("SknshRA","SknshRA","Sknsh", "Sknsh")
results<-diffExpressedVariants(countsData, conditions)
writeOutputKissDE(results, adjPvalMax = 0.05, dPSImin = 0.1, output = "kissDE-output.tab")
docker run --rm -p 80:3838 -v ~/training_Sknsh:/data dwishsan/kissplice-pipeline kissDE.R -f /data/training_Sknsh_k2rg_Type_1.tsv -c SknshRA,SknshRA,Sknsh,Sknsh -o /data/kDE_results.tsv --k2rg
The output file can be found here: kissDE-output.tabVisualising AS events of interest (optional)
Reads mapped to the annotated reference genome are available here. You can download them on your machine using wget.wget ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/STAR_readsAlign/*
Then launch IGV, select hg38 genome, import these tracks, zoom on a gene (for instance MYL6), right-click on the left panel of one of the tracks, select sashimi plot option. You should visualise something like this (you can filter out exon junctions supported by fewer than 5 reads by right-clicking and selecting "Set junction Coverage Min"):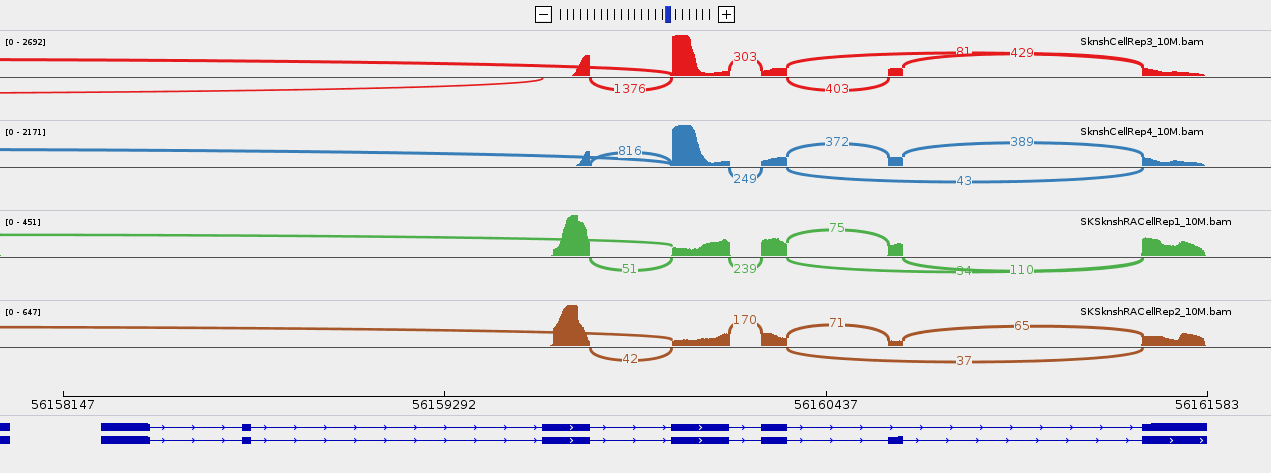
KisSplice on LCL data
The data
The subsampled fastq files can be found here. Those correspond to paired-end Illumina sequencing of polyA+ RNAs from a Tayb-Linder (TALS) patient's LCL and controls. TALS is a rare developmental syndrome in which the U4atac snRNA from the minor (or U12) spliceosome is mutated, resulting in minor splicing defects. There are two experimental conditions (control, patient), and two technicale replicates per condition. Only reads aligned to a set of predefined genes are analysed here. The genes are : KIF3A, MPHOSPH9, CCDC84, VPS11, TRPM7, EED, ACTB, TMSB10, GAPDH, EEF1A1, B2M, TMSB4X. This include between 1M and 4M reads per file. For an analysis of the full dataset, see this publication
Running KisSplice
Here is the command in order to run KisSplice on this dataset :
wget ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/data/LCL/smallDataset/*
kissplice -r P1_1_R1.fq -r P1_1_R2.fq -r P1_2_R1.fq -r P1_2_R2.fq -r C1_1_R1.fq -r C1_1_R2.fq -r C1_2_R1.fq -r C1_2_R2.fq --counts 2
mkdir ~/training_TALS
wget -P ~/training_TALS ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/data/LCL/smallDataset/*
docker run --rm -v ~/training_TALS:/data dwishsan/kissplice-pipeline kissplice -r /data/P1_1_R1.fq -r /data/P1_1_R2.fq -r /data/P1_2_R1.fq -r /data/P1_2_R2.fq -r /data/C1_1_R1.fq -r /data/C1_1_R2.fq -r /data/C1_2_R1.fq -r /data/C1_2_R2.fq --counts 2 -o /data/KisSplice
All output files of kissplice are here
Aligning to the reference genome
STAR can be used to align the alternative splicing events found by kissplice back to the reference genome. Here is the reference genome in fasta format : Homo_sapiens.GRCh38.dna_sm.primary_assembly.faAnd the associated gtf : gencode.v28.annotation.formatedChr.gtf
Building STAR index of the genome :
STAR --runMode genomeGenerate --genomeDir STARIndex_GRCh38_Gencode28 --genomeFastaFiles Homo_sapiens.GRCh38.dna_sm.primary_assembly.fa --sjdbGTFfile gencode.v28.annotation.formatedChr.gtf
The build index can be found hereAligning to the reference genome :
STARlong --genomeDir STARIndex_GRCh38_Gencode28/ --readFilesIn TALS_LCL_results_k41_coherents_type_1.fa --outFileNamePrefix TALS_LCL_Type_1_
The alignment (in sam format) can be found hereDocker can also be used to run STAR, or any other aligner, with, for instance, the Biocontainers image
Running KisSplice2RefGenome
wget -P ~/training_TALS ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/STAR_asEventAlign/LCL/TALS_LCL_Type_1_Aligned.out.sam
wget -P ~/training_TALS ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/annotation/gencode.v28.annotation.formatedChr.gtf
kissplice2refgenome -a gencode.v28.annotation.formatedChr.gtf -o TALS_LCL_k2rg_Type_1 --counts 2 --pairedEnd TALS_LCL_Type_1_Aligned.out.sam
wget -P ~/training_TALS ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/STAR_asEventAlign/LCL/TALS_LCL_Type_1_Aligned.out.sam
wget -P ~/training_TALS ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/annotation/gencode.v28.annotation.formatedChr.gtf
docker run --rm -v ~/training_TALS:/data dwishsan/kissplice-pipeline kissplice2refgenome -a /data/gencode.v28.annotation.formatedChr.gtf -o /data/TALS_LCL_k2rg_Type_1 --counts 2 --pairedEnd /data/TALS_LCL_Type_1_Aligned.out.sam
The ouput file can be found here : TALS_LCL_k2rg_Type_1.tsv
Running kissDE
The last version of kissDE can be downloaded here.
#!/usr/bin/Rscript
library(kissDE) # version 1.2 !
countsData<-kissplice2counts(fileName = "TALS_LCL_k2rg_Type_1",counts = 2,pairedEnd = T, k2rg=TRUE)
conditions<-c("P","P","C", "C")
results<-diffExpressedVariants(countsData, conditions,technicalReplicates = T)
writeOutputKissDE(results, output = "TALS_LCL_kDE_type_1")
writeOutputKissDE(results, output = "TALS_LCL_kDE_type_1_signi", adjPvalMax = 0.05)
docker run --rm -p 80:3838 -v ~/training_TALS:/data dwishsan/kissplice-pipeline kissDE.R -f /data/TALS_LCL_k2rg_Type_1.tsv -c P,P,C,C -o /data/kDE_results.tsv --pairedEnd --k2rg
The output files can be found here: kissDE outputVisualising AS events of interest (optional)
Reads mapped to the annotated reference genome are available here. You can download them on your machine using wget.wget ftp://pbil.univ-lyon1.fr/pub/logiciel/kissplice/Formation/STAR_readsAlign/LCL/*
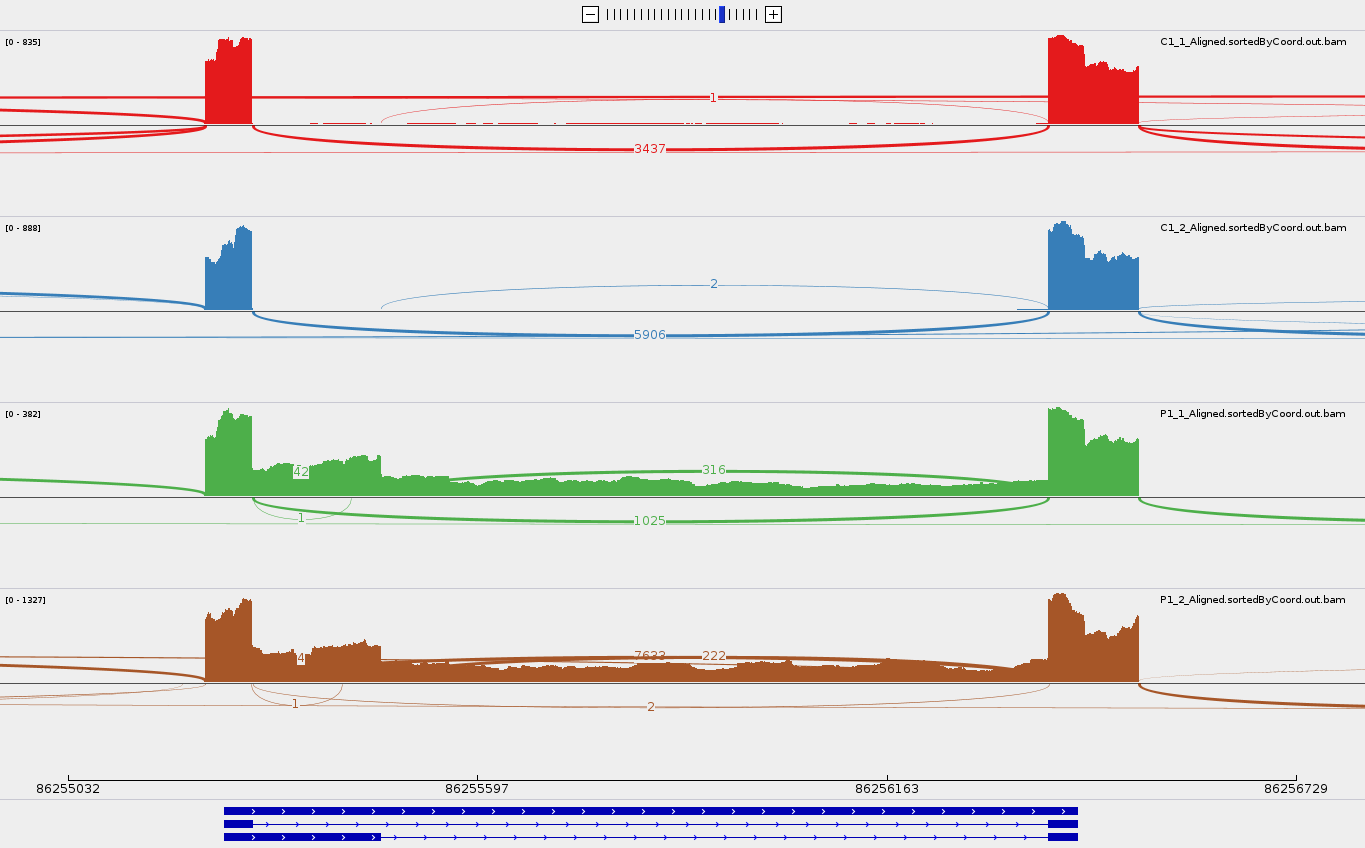
Then launch IGV, select hg38 genome, import these alignement tracks (File > Load from File), you may want to load the gtf of the significative events too, zoom on a gene (for instance EED) or a genomic position (for instance : chr11:86,254,939-86,256,824), right-click on the left panel of one of the tracks, select sashimi plot option. You should visualise something like this (you can filter out exon junctions supported by fewer than 5 reads by right-clicking and selecting "Set junction Coverage Min"):